A Genome Context Viewer (GCV) “Test-Drive”
Written by: Dr. Nevin Young
- June 25, 2019
As we see in Andrew Farmer’s post Bringing Macrosynteny to the GCV Multi-view, the Genome Context Viewer (GCV) is a web-app that provides interactive and synchronized comparative genomics visualizations. But how does it perform with real-world research questions? As part of the March, 2019 “Future of Legume Genetic Data Resources” workshop at the Noble Research Institute in Ardmore, OK, I decided to take GCV out for a test-drive to help answer the question, “Are symbiosis genes, clustered together in the Medicago truncatula genome into so-called ‘symbiotic islands’, also conserved, syntenic and co-linear in the genomes of other legume species?” Because the test-drive was so limited in scope, there’s really no way to reach definitive conclusions about symbiotic islands. But the study did provide insights into symbiotic islands, strengths and limitations of GCV – and also the challenges of comparative genomics based on sequence assemblies coming from diverse sources.
Many M. truncatula Nodule Expressed Genes Are Co-Localized in Genomic Clusters
In a recent paper in Nature Plants (4(12) 1017-1025), Pecrix et al used a substantially improved genome sequence assembly of M. truncatula to show that nearly 40% of genes upregulated in nodules or expressed in the nodulation differentiation zone colocalize in physical genomic clusters. They call these clusters, “symbiotic islands”. Of course, it’s a bit surprising that no one has seen this before in other well sequenced legume genomes. Is that because M. truncatula is somehow unique in the way its nodule-related genes are organized or are symbiosis islands found throughout legume genomes if only one takes the time to look? To answer this question, GCV seems like an useful tool.
Searching for Conserved Symbiotic Islands Using GCV
The strengths of GCV as a comparative genomics visualization tool are many. As a federated resource, users can seamlessly compare nearly every sequenced legume genome and observe directly the level of conservation – or lack thereof – surrounding any genome region of interest. Still, it’s important to remember that GCV is genic in perspective. It begins with gene calls in the compared genomes and then uses gene families (typically Phytozome-based) to infer orthology and create parallel linear displays of glyphs representing the ortholog groups. For the symbiotic island study, I chose a small number of the colocalized nodule-expressed gene clusters previously reported by Pecrix et al and then asked what they looked like across legume genomes. To get a peek at different types of symbiotic islands, one was chosen to be composed exclusively of (nodule-related) metabolic genes, another of regulatory factors, and still others,members of nodule-related small secreted peptide families (Trujillo et al 2019; Alunni et al 2007). Potentially, gene orthology and order could be fully maintained throughout an island, partially conserved, or mostly disrupted. Here, I report the results with four different symbiotic islands.
Xyloglucan Transferase Island (centered at 4g0072631) Interactive version
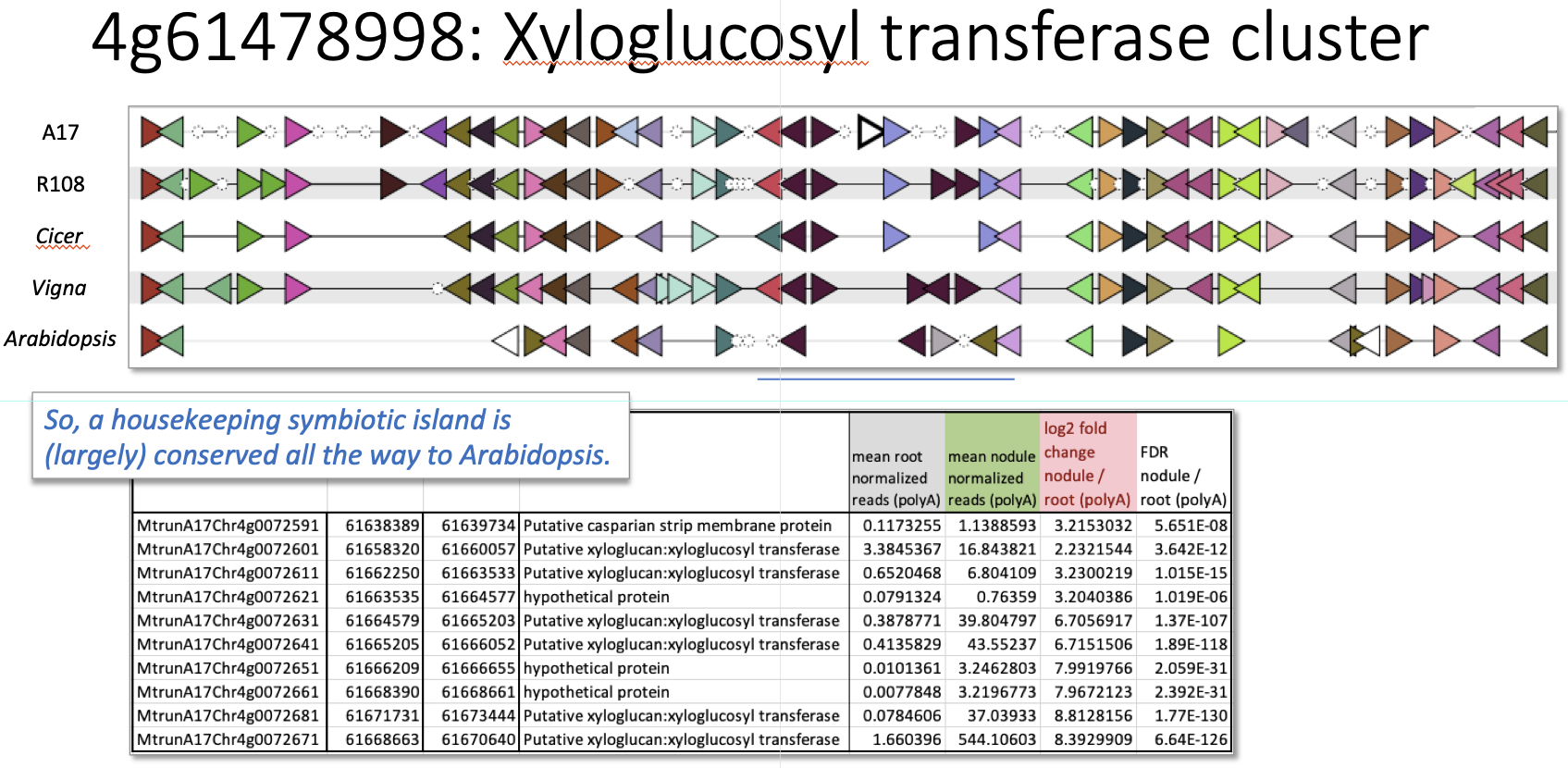
The first symbiotic island analyzed is found on chromosome 4 of M. truncatula genotype A17 and it consists of six xyloglucan transferase genes together with one gene annotated as a Casparian strip membrane protein along with two other hypothetical proteins. All of these genes exhibit a log2-fold increase of at least 2 in nodules versus roots (and FDRs < 10E-08) and all are co-localized in a region of 32 kb (the blue underline in this and other figures). GCV does an outstanding job focusing in on the target genome region (and its surroundings) and highlights the overall level of synteny and co-linearity with other legume species (in this case, a second M. truncatula genotype known as R108, Cicer, and Vigna). In this example, it’s clear the symbiotic island is, indeed, well conserved across legumes. More surprising, the same cluster of genes in basically the same order/orientation is also mostly conserved in Arabidopsis, a non-legume species incapable of symbiosis or nodulation. Apparently, this particular syntentic island may be well-conserved across angiosperms (or at least dicots), it just isn't an (exclusively) symbosis island.
Mixed Gene Family Island (centered at 6g0488461) Interactive version
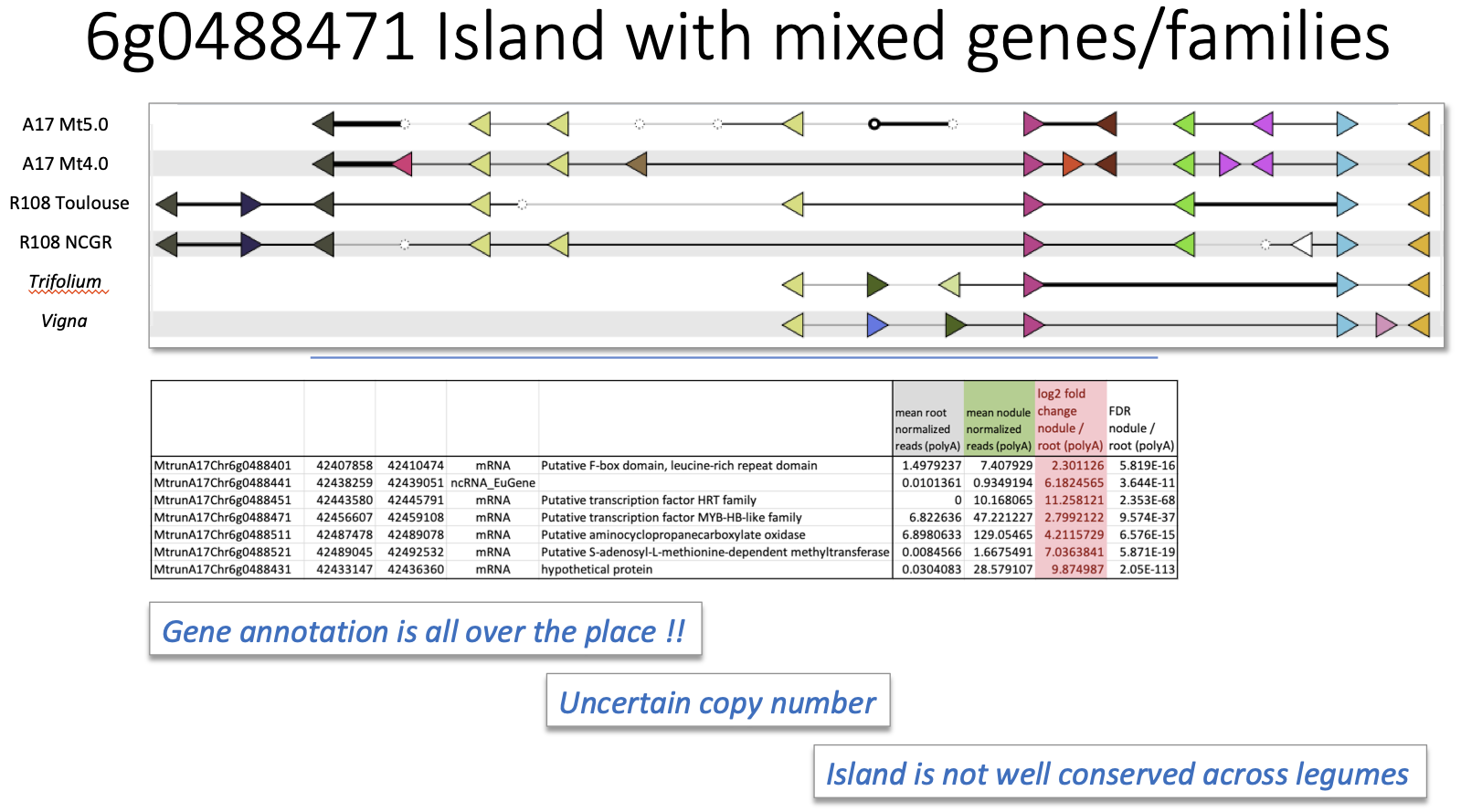
The second symbiotic island analyzed is located on chromosome 6 of M. truncatula. It consists of an F-box/LRR gene, an HRT transcription factor, a MYB transcription factor, an aminocyclopropanecarboxlate oxidase, and a hypothetical protein.The island also contains a nodule-specific non-coding RNA (a frequent sequence element in the symbiotic islands of the Pecrix et al analysis). In this case, however, GCV uncovers a region of disrupted synteny and gene order, even among different assemblies of the same M. truncatula genotype (A17) – Mt4.0 vs Mt5.0. These are two published A17 genome sequences of differing quality and differing assembly/annotation pipelines (Tang et al 2014, Pecrix et al 2018). Gene content and order is also variable in comparison with two different assemblies of M. truncatula genotype R108 reported in two recent publications (Moll et al 2017; Pecrix et al 2018). Synteny and colinearity is likewise disrupted in comparison with legume species Trifolium and Vigna. Moreover, gene copy number varies across compared genomes. Apparently, gene annotation – in addition to real gene content and gene family membership – differs across the different sequencing groups. Ultimately, GCV visualization suggests this symbiotic island is not well-conserved, even within M. truncatula, and it is likewise disrupted in comparison with other legume genomes. Nevertheless, confidence in this conclusion is limited by the variability in the way different genomes were assembled and annotated, complicating the inference of valid gene content differences among genomes.
Nodule-specific PLAT Domain (NPD) Island (centered at 2g0332661) Interactive version
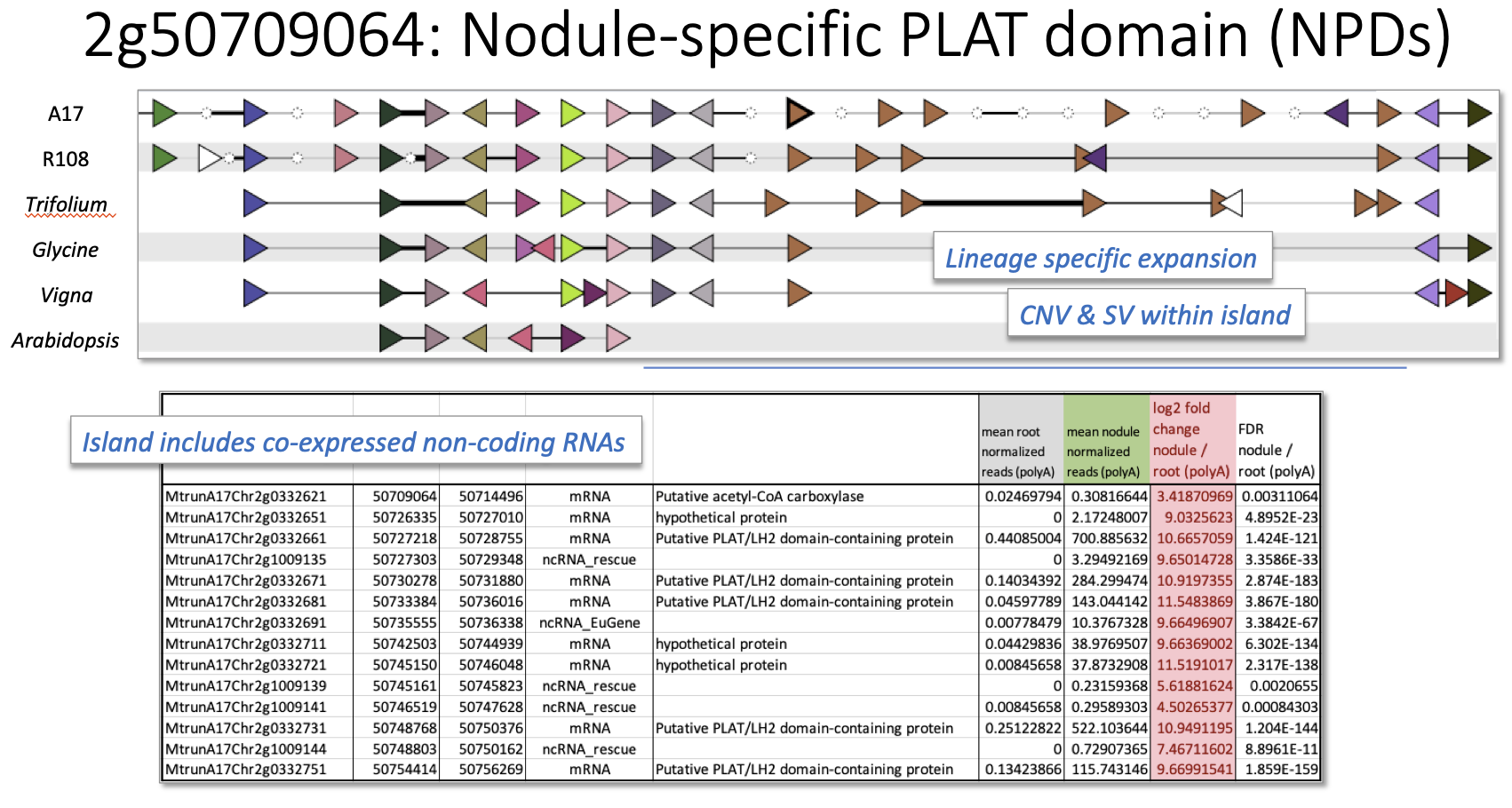
The third symbiotic island analyzed is located on chromosome 2 of M. truncatula and consists of a cluster of nodule-specific PLAT domain protein (NPD) genes (Trujillo et al 2019) together with an acetyl-CoA carboxlase, three hypothetical protein genes, and five non-coding RNAs. Here, GCV uncovers a region of reasonable synteny/colinearity, but with the NPD genes experiencing a lineage-specific expansion in M. truncatula and Trifolium (both “IRLC” legume species) in comparison with Glycine or Vigna. The target region is entirely absent from the Arabidopsis genome. Ultimately, this symbiosis island is conserved among the IRLC species and the overall region also shows some conservation with Glycine and Vigna beyond the notable copy number variation in NPD family members.
Nodule Cysteine Rich (NCR) Peptide Island (centered at 7g0216211) Interactive version
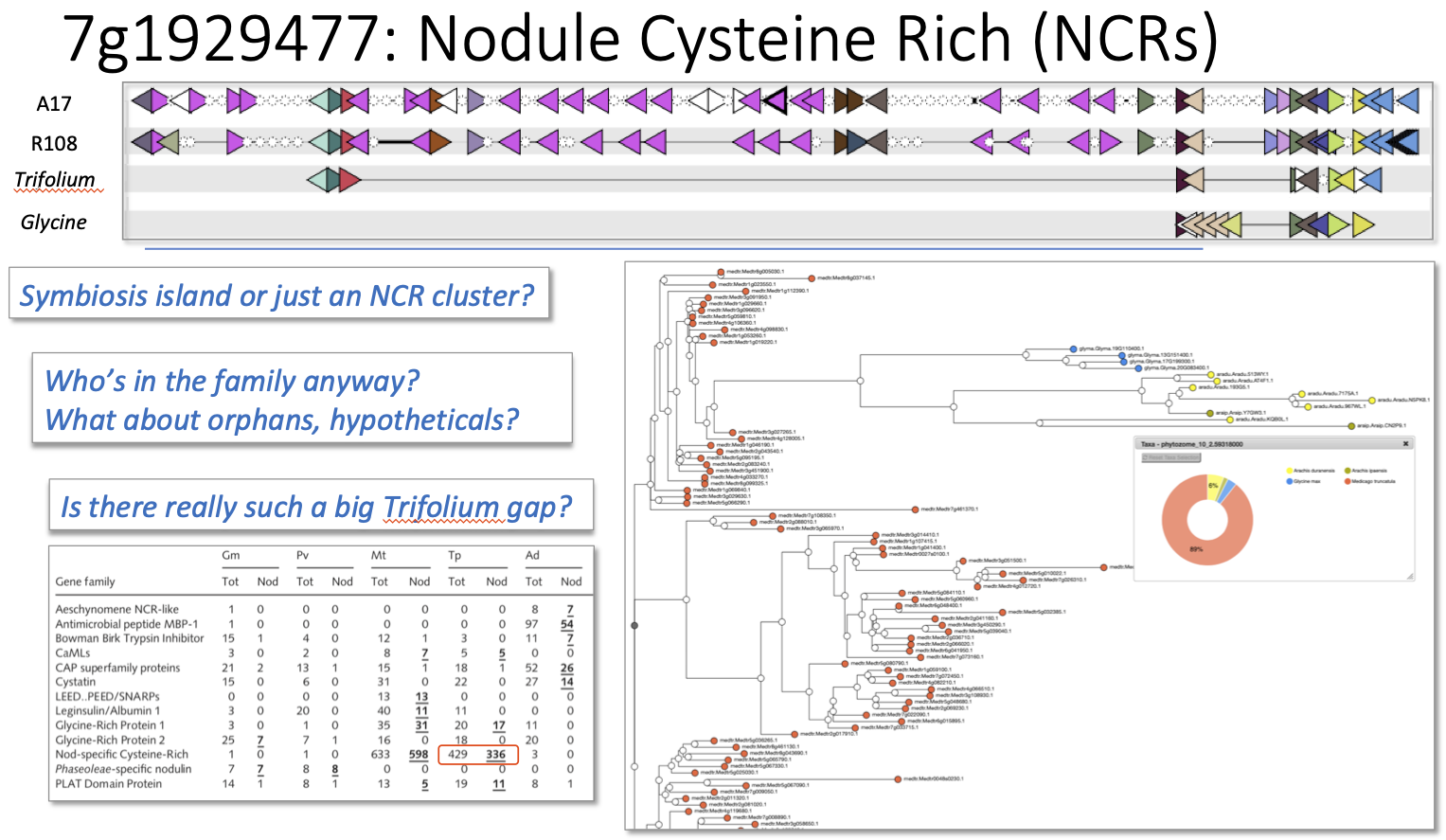
The final symbiotic island is located on chromosome 7 of M. truncatula and it differs from the others in consisting of a very large cluster of nodule-specific cysteine rich (NCR) peptides. It consists of 18 annotated NCR genes, 8 hypothetical proteins (which are probably also NCRs), and 12 ncRNAs, all in a genome region of 135 kb. Here, differing annotation across different sequencing groups makes the comparison almost impossible. GCV clearly shows conservation across the two sequenced M. truncatula genomes – though with notable differences in CNVs and sub-family membership that may be real or may just be differences in annotation and/or the parameters used to define gene sub-families. But in the comparison with Trifolium, where a few of the flanking genes do seem to be conserved, the entire NCR cluster is missing! Is this real? It can’t simply be that Trifolium lacks NCRs. Trifolium has been estimated to have 336 NCRs genome-wide (compared with 598 genome-wide in M. truncatula). But given this puzzling paradox in the defining feature of this symbiotic island, GCV leaves us with more questions than answers – and reminds us that the success of a comparative genomic analysis depends as much on the way genomes are assembled and annotated as the actual presence or absence of synteny.
Lessons from the GCV Test-Drive
This test-drive of the Genome Context Viewer demonstrates just how powerful the tool can be at observing conserved gene order and orientation. Visualizations are intuitive, information can be drawn from diverse sources, multiple genomes can be examined, comparisons can span vastly different size scales, and link-outs to other tools enable deeper analyses. CNVs, other types of SVs, as well as hypothetical and orphaned genes are handled in simple and intuitive ways. However, there are challenges. It can be difficult tracking gene families, and indeed, interpretation of genome comparisons is exceptionally sensitive to what is called a family member (or not), and this is typically performed by an external tool out of the hands of users. The factors that define syntenic regions in the first place also rely on tricky parameters that might make sense informatically, but not necessarily in a genomic sense.
Analysis of symbiotic islands using GCV demonstrates that islands display distinctly different degrees of conservation across genomes. Here, only four symbiotic islands were analyzed out of a total 270 described in Pecrix et al (2018). However, GCV analysis shows that the level of conservation differs for each one. There is one island that is highly conserved all the way to Arabidopsis, another throughout legumes, and others that are disrupted even within Medicago.
GCV analysis of symbiotic islands also illustrates another important feature (limitation?) of this type of analysis. Comparative genomics based on “federated” genome assemblies is exceedingly sensitive to the sequencing, assembly, and annotation philosophies of different research groups. When GCV displays conserved regions, one can be reasonable sure of synteny. But when conservation is disrupted by hypothetical or orphan genes, gaps, CNVs or other SVs, it is difficult to know whether one is viewing an authentic genome feature – or an informatic difference of opinion among research groups.
About the Author:

Dr. Nevin D. Young
Distinguished McKnight University Professor
University of Minnesota
Areas of Interest:
- Genomics of plants
- Symbiosis
- Disease resistance
